Drought Stress Reshapes Secondary Metabolism in Safflower (Carthamus tinctorius L.): Insights from GC–MS Metabolomics and Bioinformatic Analysis
DOI:
https://doi.org/10.64062/JPGMB.Vol2.Issue1.6Keywords:
Drought Stress; Secondary Metabolites; Safflower; GC Analysis; OctahydrocurcuminAbstract
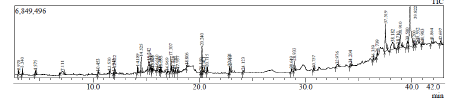
Abiotic stress–induced secondary metabolism is a key determinant of plant survival under adverse environments, and Carthamus tinctorius L. (safflower), a drought-resilient oilseed and medicinal crop, exhibits a strong capacity to accumulate bioactive metabolites under stress. In this study, GC–MS profiling of methanolic leaf extracts was integrated with bioinformatic analyses to elucidate drought-responsive secondary metabolism in safflower. GC–MS analysis revealed a diverse array of stress-induced metabolites, including phenolic acids, flavonoids, fatty acids, sterols, and terpenoids, with the notable accumulation of octahydrocurcumin, a potent antioxidant. The enrichment of phenolic and lipid-derived compounds under drought stress indicates the activation of antioxidant defense mechanisms and redox homeostasis pathways. To establish a molecular link to the observed metabolomic changes, the NCBI safflower sequence database was searched for transcripts showing homology associated with octahydrocurcumin-related stress responses. This analysis identified a drought-responsive transcript, GW584119, as a candidate sequence. The authors had previously constructed a drought-responsive cDNA library from safflower, and the nucleotide sequence corresponding to this transcript was submitted to the NCBI database under accession number GW584119. Bioinformatic characterization revealed significant sequence homology associated with octahydrocurcumin-related pathways. Comparative sequence analysis further showed partial homology with a corresponding gene in the model crop Arabidopsis thaliana (AT1G31020). Protein–protein interaction and subcellular localization analyses suggested the involvement of the encoded protein in stress-related metabolic networks, while expression profiling of AT1G31020 confirmed stress-responsive expression patterns. Overall, the integrated metabolomic and computational evidence establishes a direct link between drought stress, octahydrocurcumin accumulation, and gene-level regulation in safflower, highlighting octahydrocurcumin as a promising biochemical and molecular marker for abiotic stress tolerance and a potential target for the development of stress-resilient crops.