Genetic Markers for Predicting Cancer Immunotherapy Response
DOI:
https://doi.org/10.64062/JPGMB.Vol1.Issue4.8Keywords:
Cancer immunotherapy, Genetic markers, Animal models, Immune, checkpoint inhibitors, Preclinical, trials, Personalised, medicine, Tumour, microenvironment, Biomarker prediction; CRISPR; T-cell infiltrationAbstract
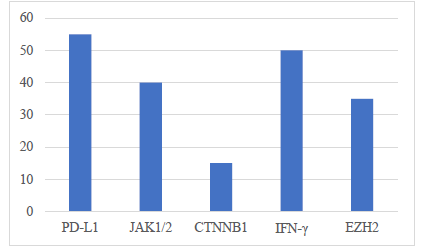
Immune checkpoint blockade and cancer immunotherapy have revised the cancer treatment therapeutic environment, but patient reactions are excessively difference. The targeted population in this systematic review is animal trials in order to understand any genetic markers effecting the efficacy of the immunotherapy. The research tries using murine models like syngeneic, genetic engineered mice and humanized mice to define key genes that are determinants of therapeutic response (PD-L1, IFN-gamma, JAK1/2, CTNNB1 and EZH2). High expression of PD-L1 and IFN-γ reported positive correlation with response and survival whereas mutation in JAK1/2 and CTNNB1 activation reported resistance because of ineffective immune infiltration and stimulation events. Inhibition of EZH2 increased immune activity partly which indicates the role of epigenetic control. The predictive value of these markers was confirmed by the analysis of data via ANOVA, Kaplan-Meier survival curves, correlation, and bioinformatics tools. Animal models will continue to be essential to mechanisms, despite species-related and methodological limitation. These results confirm the use of genetic profiling in personalized immunotherapy strategies and point to the importance of additional translational data in models more humanized as well as multi-omics.